Simulate population dynamics for one or more
species defined by dynamics objects.
Arguments
- object
a
dynamicsobject created withdynamicsor from a subsequent call tomultispeciesormetapopulation. Alternatively,objectcan be the output of a call tosimulatein the case ofsummary- nsim
the number of replicate simulations (default = 1)
- seed
optional seed used prior to initialisation and simulation to give reproducible results
- ...
ignored; included for consistency with
simulategeneric method- init
an array of initial conditions with one row per replicate and one column per population stage. If
objhas been created withmultispecies, initial conditions can be provided as a list or array with one element or slice per species, or as a matrix, in which case all species are assumed to share the same initial conditions. Defaults toNULL, in which case initial conditions are generated randomly according tooptions()$aae.pop_initialisation- options
a named
listof simulation options. Currently accepted values are:-
ntimethe number of time steps to simulate, ignored ifobjincludes acovariates(default = 50)-
keep_sliceslogicaldefining whether to keep intermediate population abundances or (ifFALSE) to return only the final time slice-
tidy_abundancesa function to handle predicted abundance data that may be non-integer. Defaults toidentity; suggested alternatives arefloor,round, orceiling-
initialise_argsa list of arguments passed to the function used to initialise abundance trajectories. Only used ifinit = NULL. Defaults tooptions()$aae.pop_lambda, which specifies lambda for Poisson random draws. The default initialisation function is defined byoptions()$aae.pop_initialisation.-
updatea function to update abundances from one time step to the next. Defaults tooptions()$aae.pop_update.- args
named list of lists passing arguments to processes defined in
object, includinginteractionformultispeciesobjects. Lists (up to one per process) can contain a mix of static, dynamic, and function arguments. Dynamic arguments must be lists with one element per time step. Function arguments must be functions that calculate arguments dynamically in each generation based on from the population dynamics object, population abundances, and time step in each generation. All other classes (e.g., single values, matrices, data frames) are treated as static arguments. Covariates contained in numeric vectors, matrices, or data frames can be formatted as dynamic arguments with theformat_covariatesfunction.argsformultispeciesobjects must have one element per species (defaults will expand automatically if not provided)- .future
flag to determine whether the future package should be used to manage updates for multispecies models (an embarrassingly parallel problem)
- x
an object to pass to
is.simulationoris.simulation.list
Value
simulation object containing replicate simulations from
a matrix population model. plot and subset methods are
defined for simulation objects
Examples
# define a population matrix (columns move to rows)
nclass <- 5
popmat <- matrix(0, nrow = nclass, ncol = nclass)
popmat[reproduction(popmat, dims = 4:5)] <- c(10, 20)
popmat[transition(popmat)] <- c(0.25, 0.3, 0.5, 0.65)
# can extract standard population matrix summary stats
lambda <- Re(eigen(popmat)$values[1])
# define a dynamics object
dyn <- dynamics(popmat)
# simulate from this (50 time steps, 100 replicates)
sims <- simulate(dyn, nsim = 100, options = list(ntime = 50))
# plot the simulated trajectories
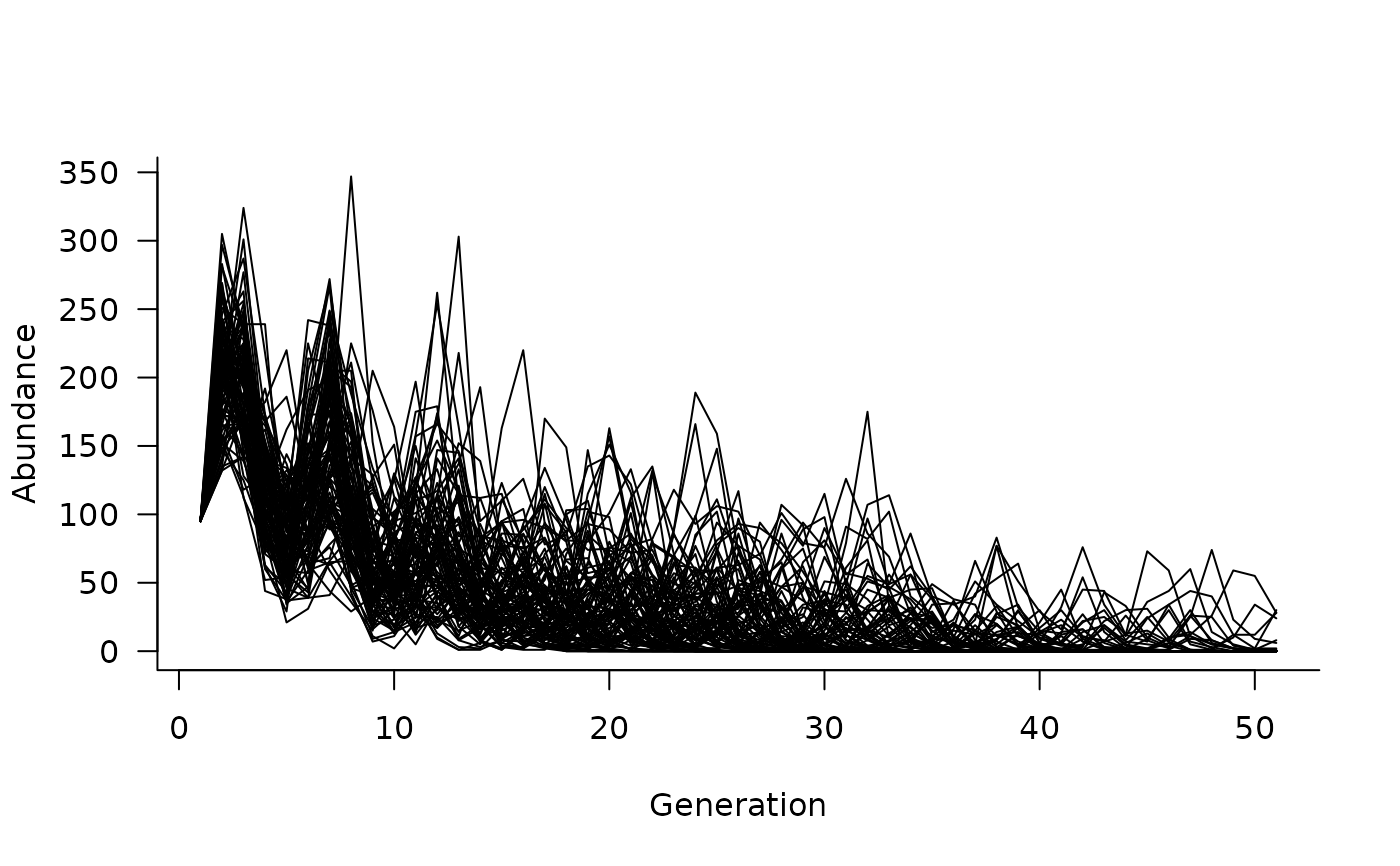
plot(sims)
 # add some density dependence
dd <- density_dependence(
masks = reproduction(popmat, dims = 4:5),
funs = ricker(1000)
)
# update the dynamics object
dyn <- update(dyn, dd)
# simulate again
sims <- simulate(dyn, nsim = 100, options = list(ntime = 50))
# and plot
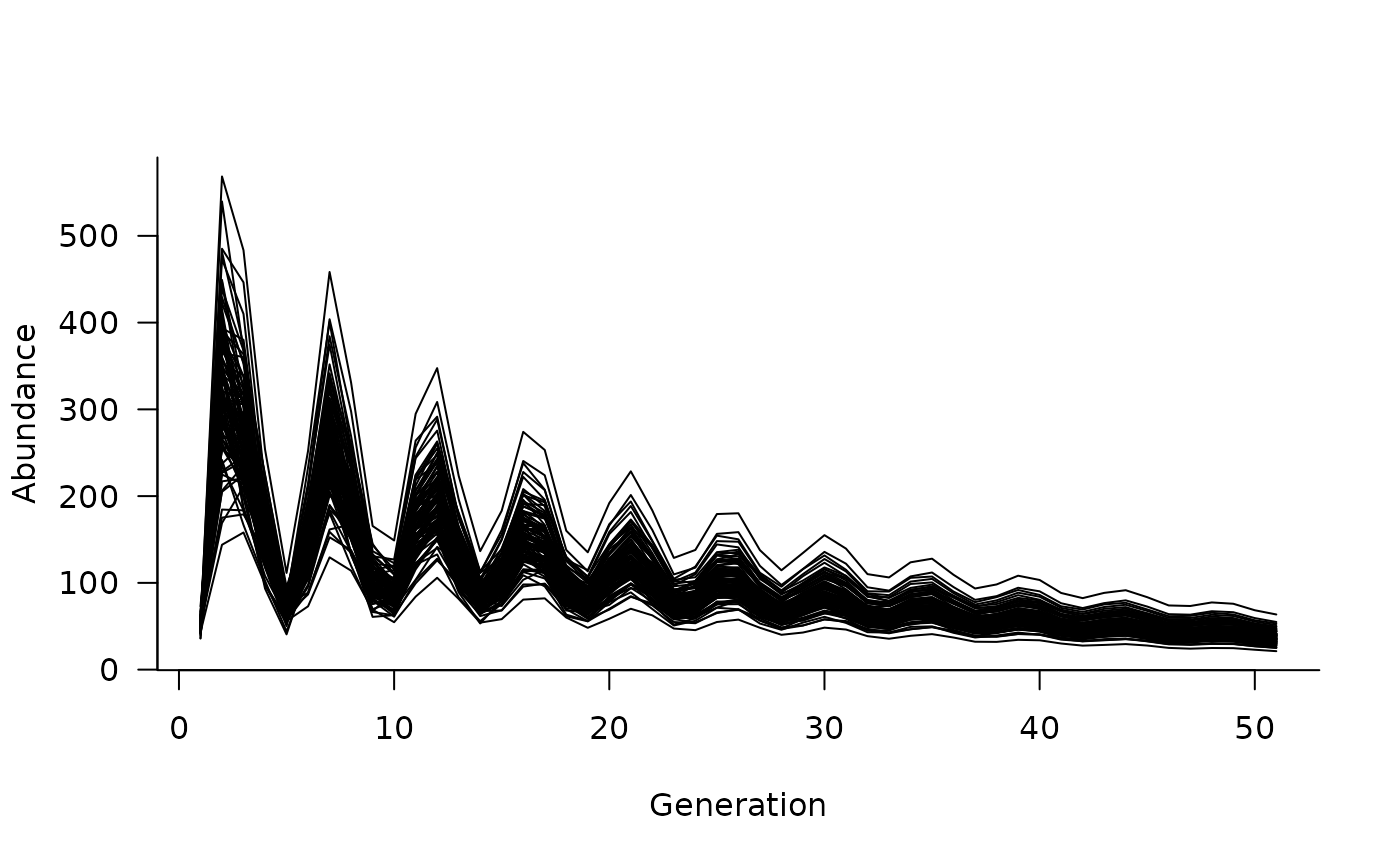
plot(sims)
# add some density dependence
dd <- density_dependence(
masks = reproduction(popmat, dims = 4:5),
funs = ricker(1000)
)
# update the dynamics object
dyn <- update(dyn, dd)
# simulate again
sims <- simulate(dyn, nsim = 100, options = list(ntime = 50))
# and plot
plot(sims)
 # what if we want to add initial conditions?
sims <- simulate(
dyn,
init = c(50, 20, 10, 10, 5),
nsim = 100,
options = list(ntime = 50),
)
# and plot again
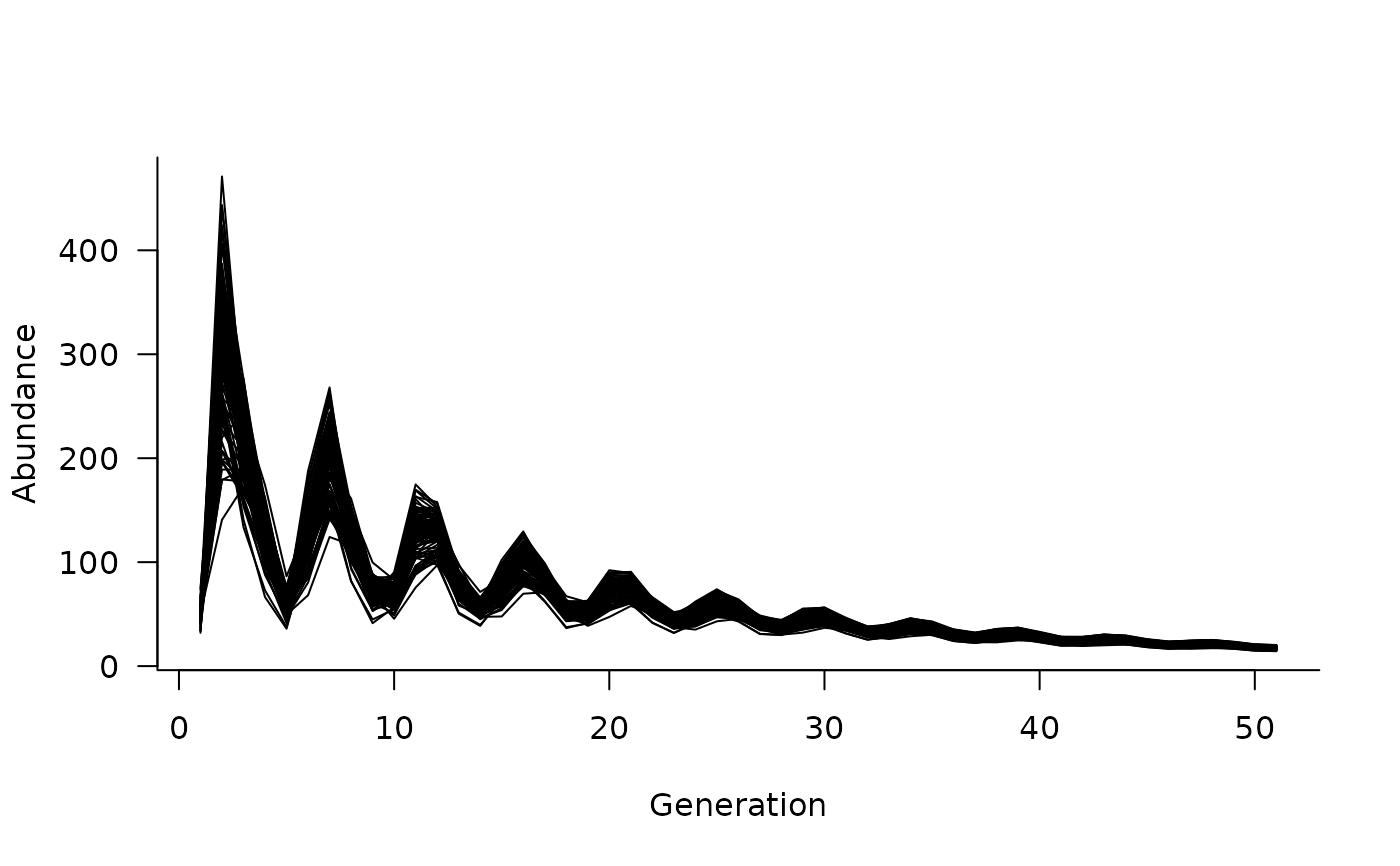
plot(sims)
# what if we want to add initial conditions?
sims <- simulate(
dyn,
init = c(50, 20, 10, 10, 5),
nsim = 100,
options = list(ntime = 50),
)
# and plot again
plot(sims)
 # note that there is only one trajectory now because
# this simulation is deterministic.
#
# let's change that by adding some environmental stochasticity
envstoch <- environmental_stochasticity(
masks = list(
reproduction(popmat, dims = 4:5),
transition(popmat)
),
funs = list(
\(x) rpois(n = length(x), lambda = x),
\(x) runif(n = length(x), min = 0.9 * x, max = pmin(1.1 * x, 1))
)
)
# update the dynamics object and simulate from it
dyn <- update(dyn, envstoch)
sims <- simulate(
dyn,
init = c(50, 20, 10, 10, 5),
nsim = 100,
options = list(ntime = 50),
)
# can also add covariates that influence vital rates
# e.g., a logistic function
covars <- covariates(
masks = transition(popmat),
funs = \(mat, x) mat * (1 / (1 + exp(-10 * x)))
)
# simulate 50 random covariate values
xvals <- matrix(runif(50), ncol = 1)
# update the dynamics object and simulate from it.
# Note that ntime is now captured in the 50 values
# of xvals, assuming we pass xvals as an argument
# to the covariates functions
dyn <- update(dyn, covars)
sims <- simulate(
dyn,
init = c(50, 20, 10, 10, 5),
nsim = 100,
args = list(covariates = format_covariates(xvals))
)
# a simple way to add demographic stochasticity is to change
# the "updater" that converts the population at time t
# to its value at time t + 1. The default in aae.pop
# uses matrix multiplication of the vital rates matrix
# and current population. A simple tweak is to update
# with binomial draws. Note that this also requires a
# change to the "tidy_abundances" option so that population
# abundances are always integer values.
sims <- simulate(
dyn,
init = c(50, 20, 10, 10, 5),
nsim = 100,
options = list(
update = update_binomial_leslie,
tidy_abundances = floor
),
args = list(covariates = format_covariates(xvals))
)
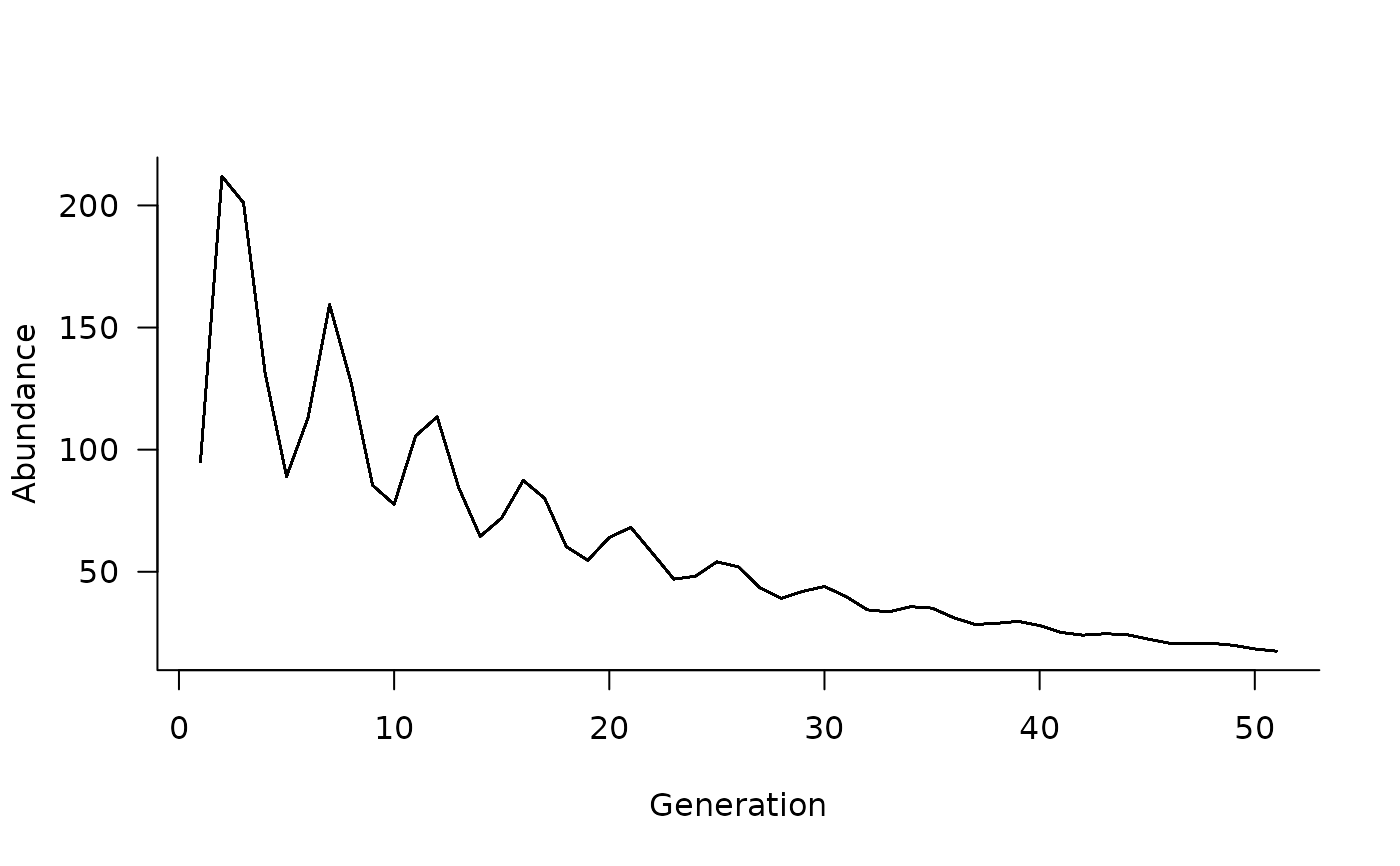
# and can plot these again
plot(sims)
# note that there is only one trajectory now because
# this simulation is deterministic.
#
# let's change that by adding some environmental stochasticity
envstoch <- environmental_stochasticity(
masks = list(
reproduction(popmat, dims = 4:5),
transition(popmat)
),
funs = list(
\(x) rpois(n = length(x), lambda = x),
\(x) runif(n = length(x), min = 0.9 * x, max = pmin(1.1 * x, 1))
)
)
# update the dynamics object and simulate from it
dyn <- update(dyn, envstoch)
sims <- simulate(
dyn,
init = c(50, 20, 10, 10, 5),
nsim = 100,
options = list(ntime = 50),
)
# can also add covariates that influence vital rates
# e.g., a logistic function
covars <- covariates(
masks = transition(popmat),
funs = \(mat, x) mat * (1 / (1 + exp(-10 * x)))
)
# simulate 50 random covariate values
xvals <- matrix(runif(50), ncol = 1)
# update the dynamics object and simulate from it.
# Note that ntime is now captured in the 50 values
# of xvals, assuming we pass xvals as an argument
# to the covariates functions
dyn <- update(dyn, covars)
sims <- simulate(
dyn,
init = c(50, 20, 10, 10, 5),
nsim = 100,
args = list(covariates = format_covariates(xvals))
)
# a simple way to add demographic stochasticity is to change
# the "updater" that converts the population at time t
# to its value at time t + 1. The default in aae.pop
# uses matrix multiplication of the vital rates matrix
# and current population. A simple tweak is to update
# with binomial draws. Note that this also requires a
# change to the "tidy_abundances" option so that population
# abundances are always integer values.
sims <- simulate(
dyn,
init = c(50, 20, 10, 10, 5),
nsim = 100,
options = list(
update = update_binomial_leslie,
tidy_abundances = floor
),
args = list(covariates = format_covariates(xvals))
)
# and can plot these again
plot(sims)